Hogwash is an R package to perform microbial genome-wide association studies (mGWAS). It work by identifying the co-convergence of genotypes and phenotypes more than would be expected by chance. Hogwash is capable of working with discrete (e.g. antibiotic resistance) and continuous (e.g. gene expression) phenotypes. A unique aspect of hogwash is its grouping feature, which allows for testing of functional units (e.g. genes, pathways) in a single test by considering emergence of any variant in the unit during an association test.
Cognac is an R package to make concatenated alignments for large genomic datasets. Important aspects of cognac are its speed, ability to handle massive genomic data sets and ability to work with genomic datasets varying in diversity (e.g. within or between species).
Crawford RD, Snitkin ES. cognac: rapid generation of concatenated gene alignments for phylogenetic inference from large, bacterial whole genome sequencing datasets. BMC Bioinformatics. 2021 Feb 15;22(1):70. PMCID: PMC7885345
Regentrans is an R package that aggregates a lot of basic functionality that our group has found useful for studying regional transmission of bacterial pathogens.
Hoffman S, Lapp Z, Wang J and Snitkin ES. regentrans: a framework and R package for using genomics to study regional pathogen transmission. medrxiv, 2021.
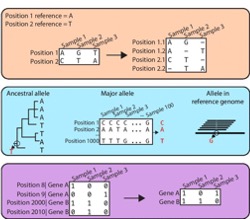
prewas is an R package that transforms genomic variants (e.g. vcf) into a binary input matrix that can feed into tools for genome-wide association. Useful aspects of prewas are the ability to re-reference variants using different approaches, handle multiple alleles at a given position and group variants by gene/pathway.
Saund K*, Lapp Z*, Thiede SN*, Pirani A, Snitkin ES. prewas: data pre-processing for more informative bacterial GWAS. Microb Genom. 2020 May;6(5)Microb Genom. 2020 May;6(5)